In Press/Submitted
 Implicit Solvation Using the Superposition Approximation (IS-SPA): Extension to Polar Solutes in Chloroform.
Implicit Solvation Using the Superposition Approximation (IS-SPA): Extension to Polar Solutes in Chloroform.
P. T. Lake, M. A. Mattson, M. McCullagh (2020)
Preprint prior to publication.
Published
 Using molecular simulations to investigate how intermolecular interactions dictate liquid structure.
Using molecular simulations to investigate how intermolecular interactions dictate liquid structure.
P. T. Lake, M. McCullagh (2020)
Chapter 3 in "Intra- and Intermolecular Interactions Between Non-covalently Bonded Species", Elsevier.
 A Hyperactive Kunjin Virus NS3 Helicase Mutant Demonstrates Increased Dissemination and Mortality in Mosquitoes.
A Hyperactive Kunjin Virus NS3 Helicase Mutant Demonstrates Increased Dissemination and Mortality in Mosquitoes.
K. E. Du Pont, N. R. Sexton, M. McCullagh, G. D. Ebel, B. J. Geiss (2020)
J. Virol., 94, e01021-20.
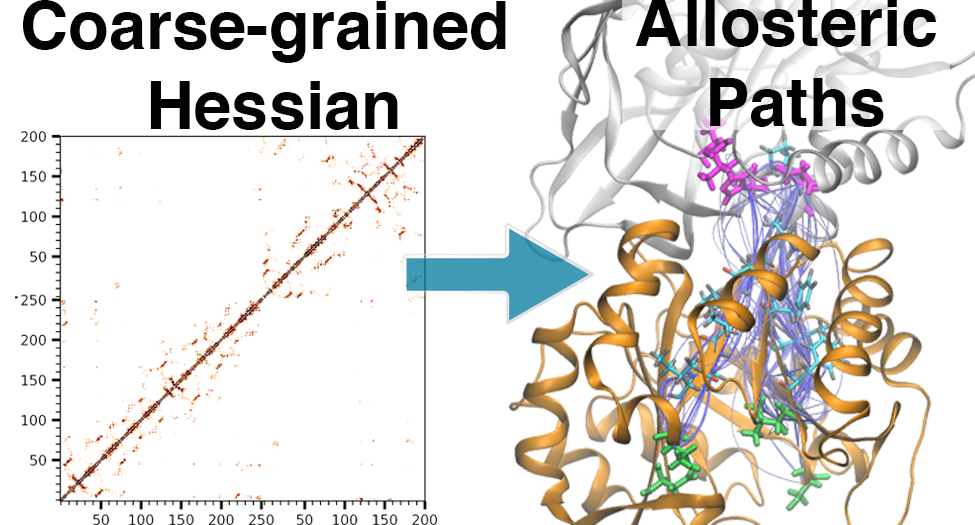
 Residue-Level Allostery Propagates Through the Effective Coarse-Grained Hessian
Residue-Level Allostery Propagates Through the Effective Coarse-Grained Hessian
P. T. Lake, R. B. Davidson, G. M. Hockey, H. Klem, M. McCullagh (2020)
J. Chem. Theory Comput., 16, 5, 3385-3395.
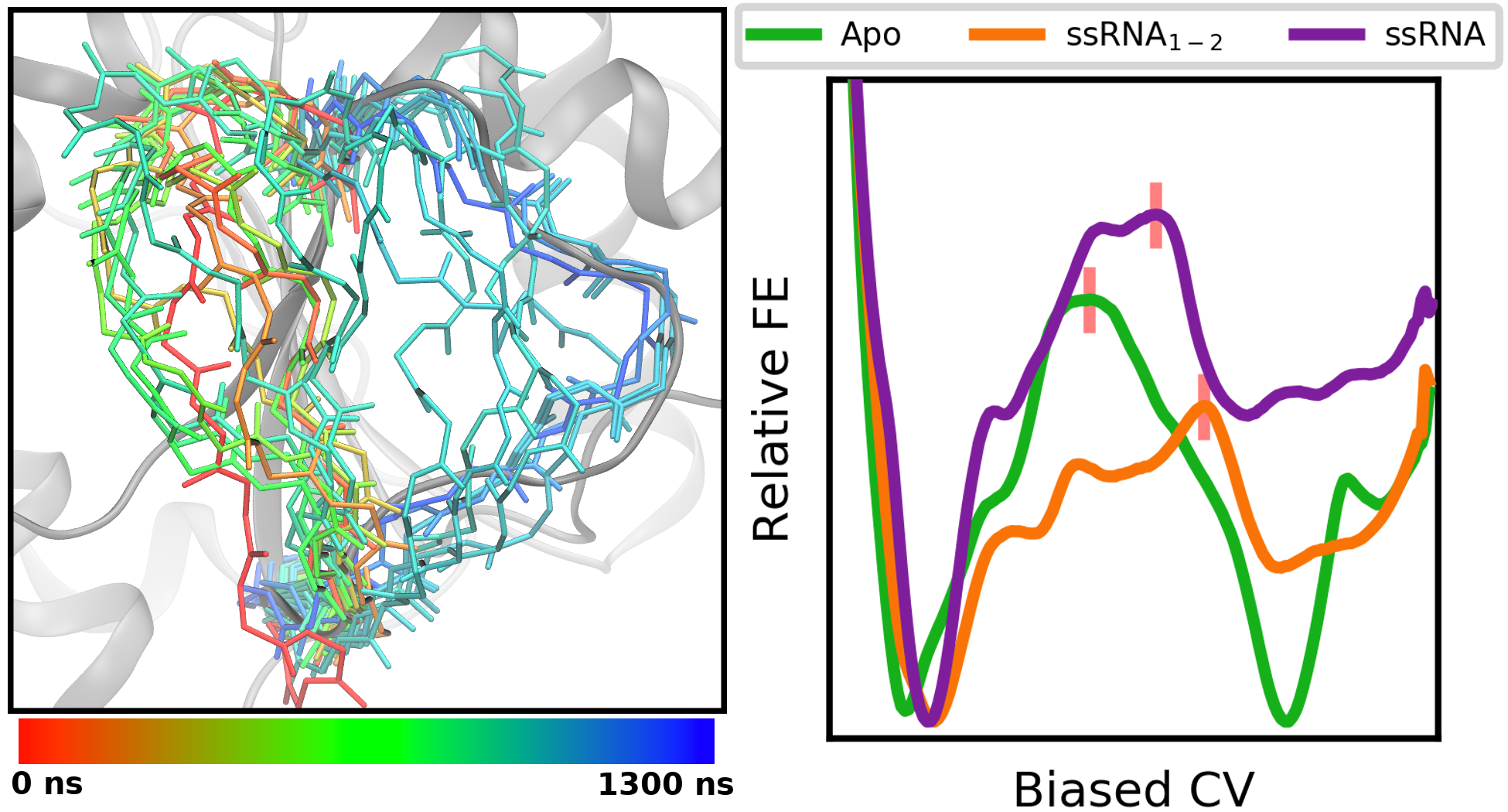 RNA-dependent structures of the RNA-binding loop in the flavivirus NS3 helicase
RNA-dependent structures of the RNA-binding loop in the flavivirus NS3 helicase
R. B. Davidson, J, Hendrix, B. J. Geiss, M. McCullagh (2020)
J. Phys. Chem. B, 124, 12, 2371-2381
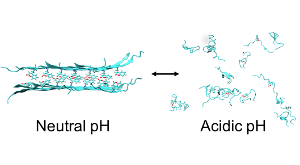 The Role of Hydrophobicity in the Stability and pH-Switchability of (RXDX)4 and Coumarin-(RXDX)4 Conjugate β-Sheets
The Role of Hydrophobicity in the Stability and pH-Switchability of (RXDX)4 and Coumarin-(RXDX)4 Conjugate β-Sheets
R. Weber, M. McCullagh (2020)
J. Phys. Chem. B, 124, 9, 1723-1732
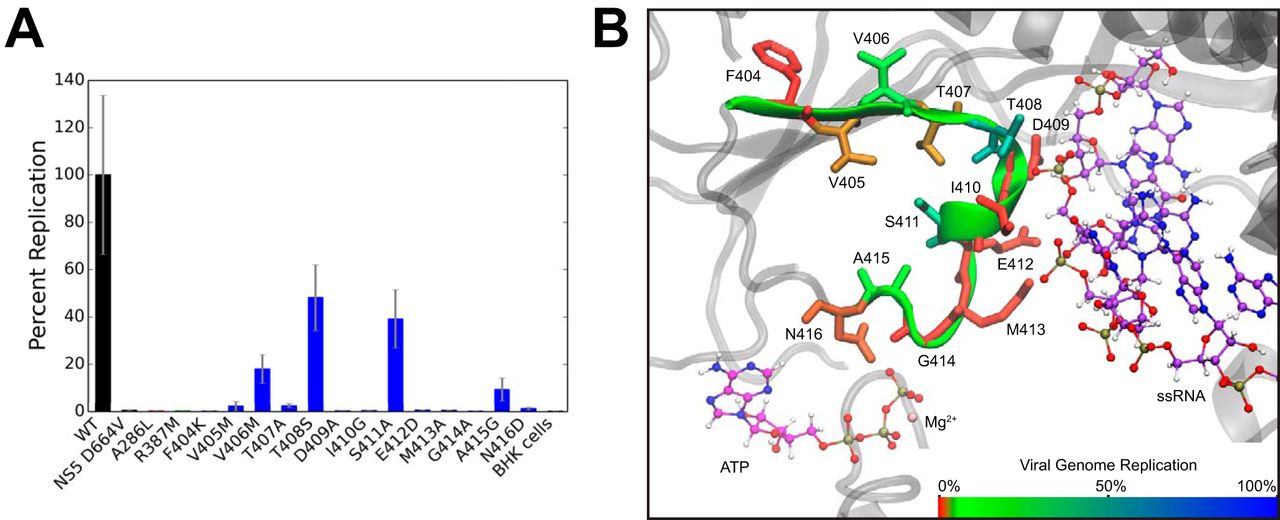
 Motif V regulates energy transduction between the flavivirus NS3 ATPase and RNA-binding cleft
Motif V regulates energy transduction between the flavivirus NS3 ATPase and RNA-binding cleft
K. E. Du Pont, R. B. Davidson, M. McCullagh, B. J. Geiss (2020)
J. Biol. Chem., 295, 6, 1551-1564
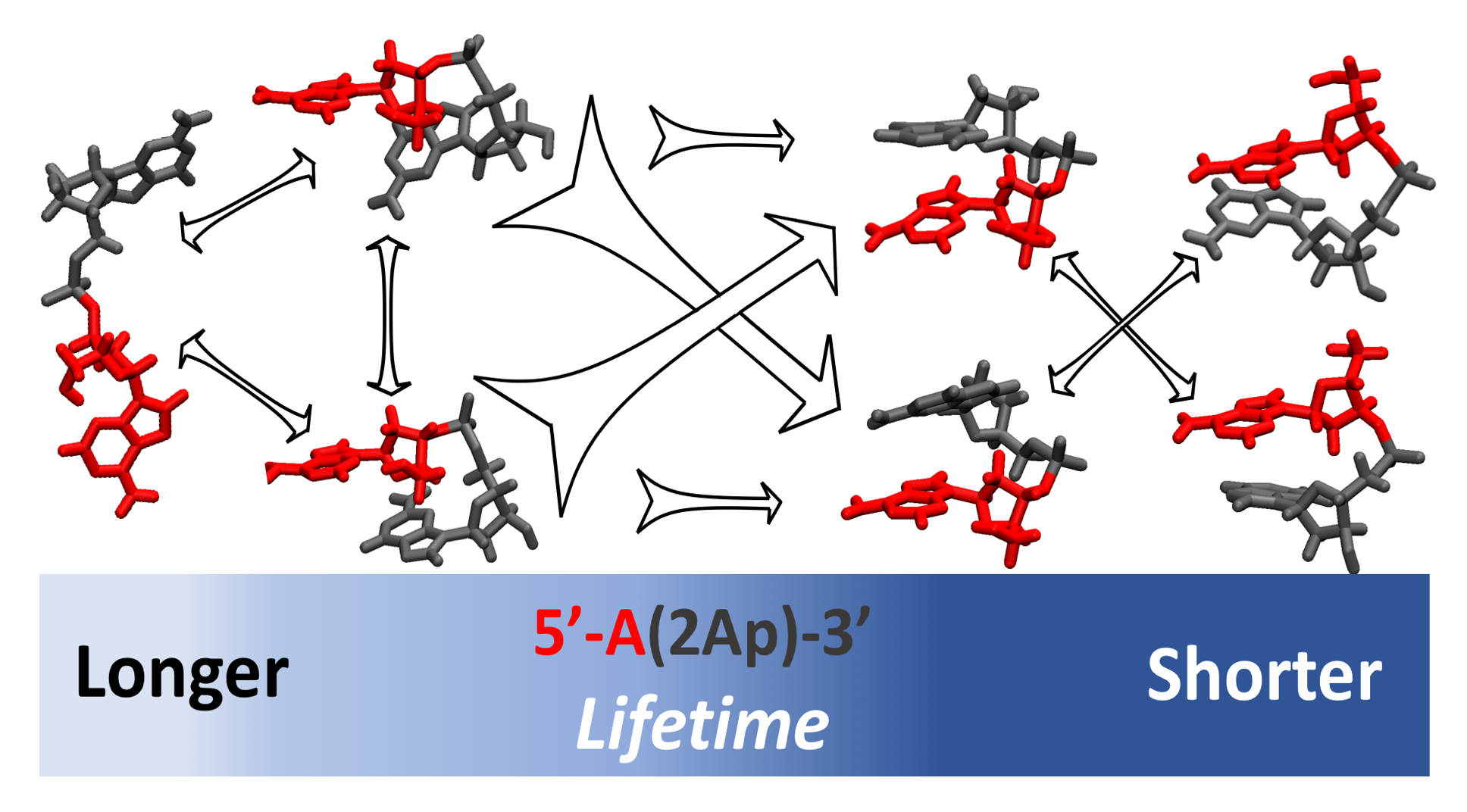 Molecular Dynamics Simulations of 2-Aminopurine-Labeled Dinucleoside Monophosphates Reveal Multiscale Stacking Kinetics
Molecular Dynamics Simulations of 2-Aminopurine-Labeled Dinucleoside Monophosphates Reveal Multiscale Stacking Kinetics
J. M. Remington, M. McCullagh, B. Kohler (2019)
J. Phys. Chem. B, 123, 10, 2291-2304
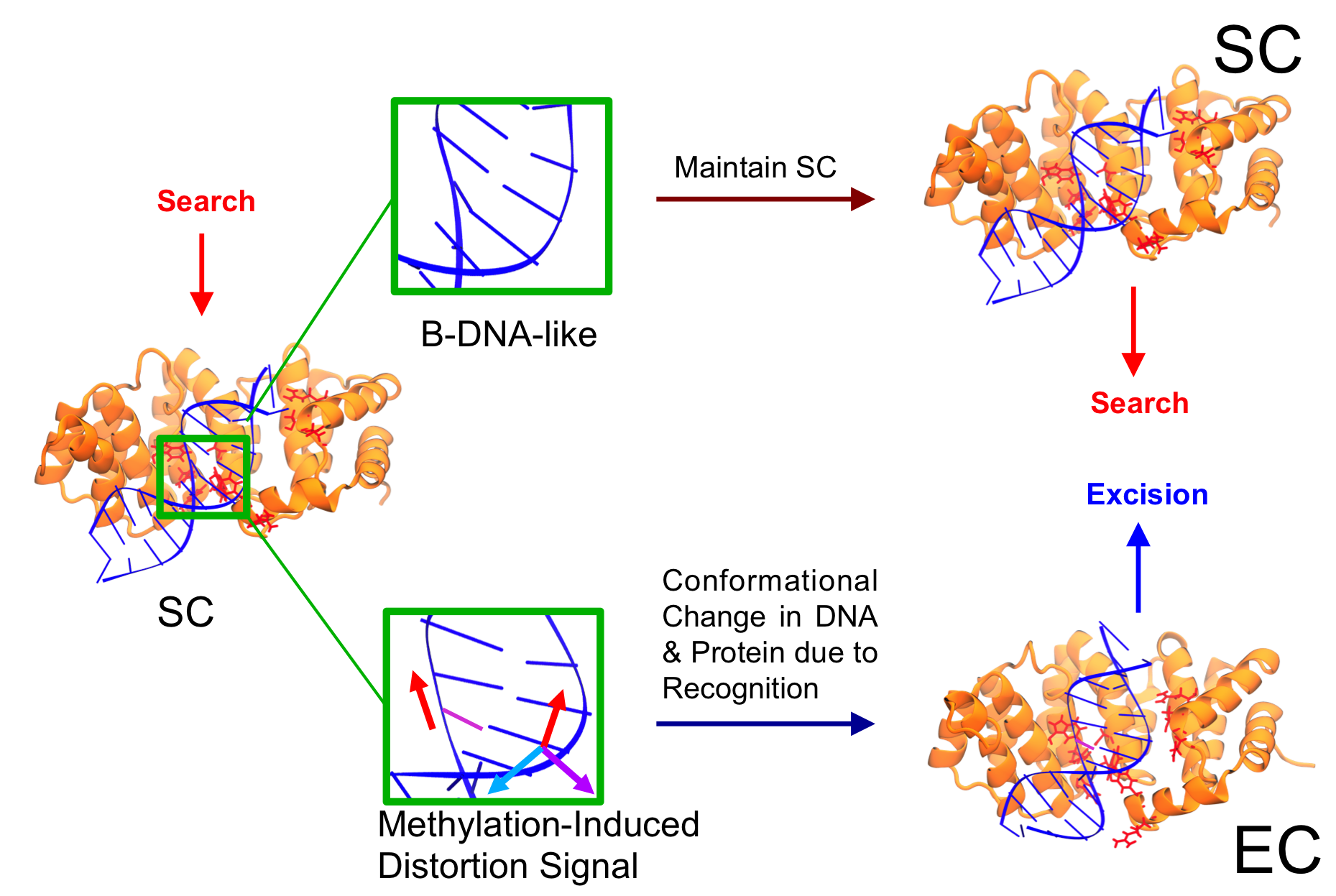
Characterization of the Search Complex and Recognition Mechanism of the AlkD DNA Glycosylase
K. A. Votaw, M. McCullagh (2019)
J. Phys. Chem. B, 123, 1, 95-105
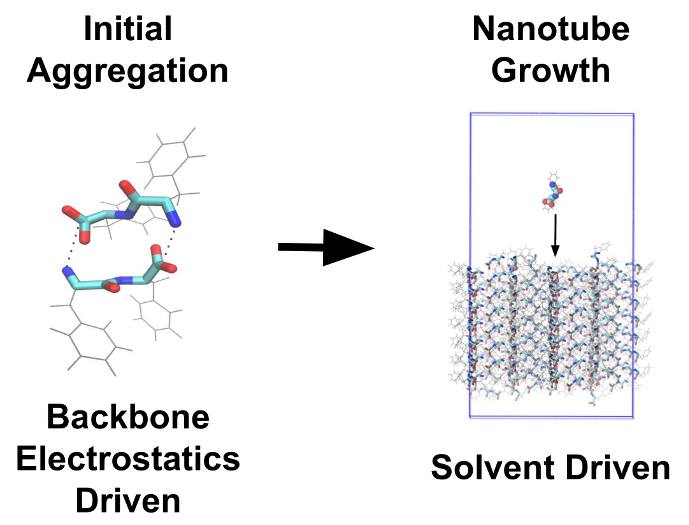 The Initial Aggregation and Ordering Mechanism of Diphenylalanine From Microsecond All-Atom Molecular Dynamics Simulations
The Initial Aggregation and Ordering Mechanism of Diphenylalanine From Microsecond All-Atom Molecular Dynamics Simulations
J. Anderson, P. T. Lake, M. McCullagh (2018)
J. Phys. Chem. B, 122, 51, 12331-12341
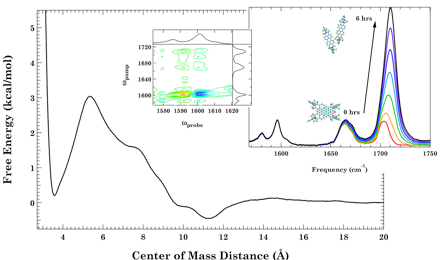 Elucidating Structural Evolution of Perylene Diimide Aggregates Using Vibrational Spectroscopy and Molecular Dynamics Simulations
Elucidating Structural Evolution of Perylene Diimide Aggregates Using Vibrational Spectroscopy and Molecular Dynamics Simulations
M. Mattson, T. Green, P. T. Lake, M. McCullagh, A. Krummel (2018)
J. Phys. Chem. B, 122, 18, 4891-4900
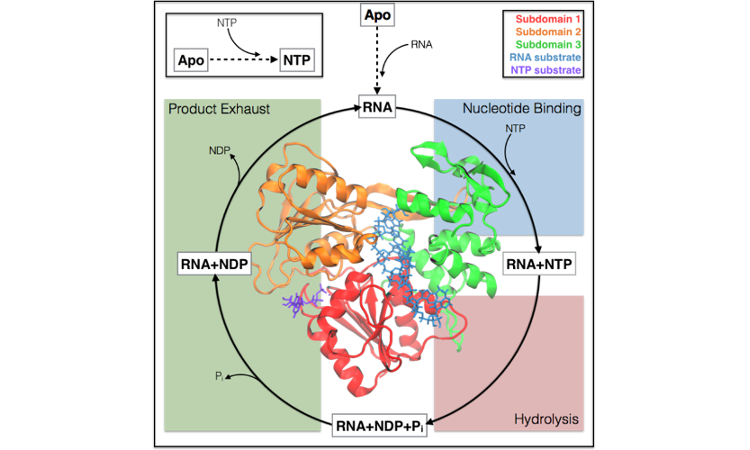 Allostery in the dengue virus NS3 helicase: Insights into the NTPase cycle from molecular simulations
Allostery in the dengue virus NS3 helicase: Insights into the NTPase cycle from molecular simulations
R. B. Davidson, J. Hendrix, B. J. Geiss, M. McCullagh (2018)
PLOS Comp. Biol., 14, 4, e1006103
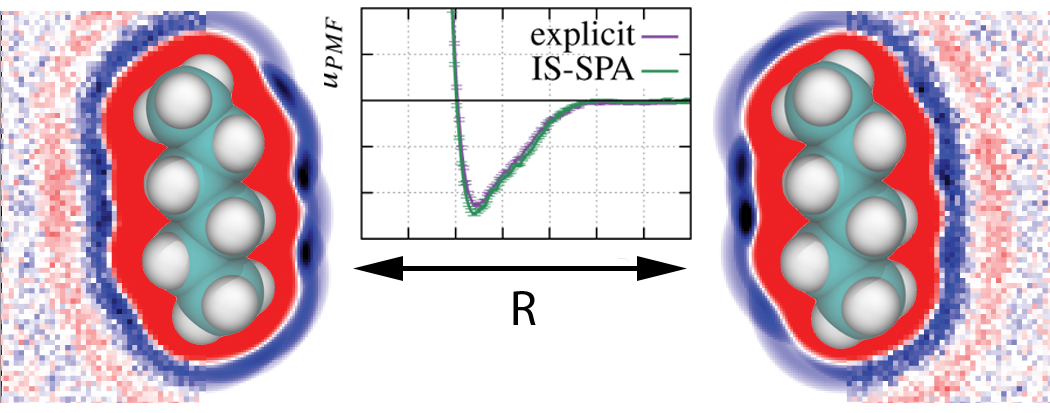 Implicit Solvation Using the Superposition Approximation (IS-SPA): An Implicit Treatment of the Nonpolar Component to Solvation for Simulating Molecular Aggregation
Implicit Solvation Using the Superposition Approximation (IS-SPA): An Implicit Treatment of the Nonpolar Component to Solvation for Simulating Molecular Aggregation
P. T. Lake, M. McCullagh (2017)
J. Chem. Theory Comput., 13, 12, 5911-5924
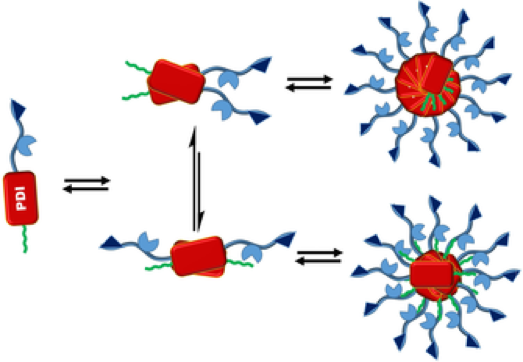 Self-Assembly of Perylenediimide-Single-Stand-DNA Conjugates: Employing Hydrophobic Interactions and DNA Base-Pairing To Create a Diverse Structural Space
Self-Assembly of Perylenediimide-Single-Stand-DNA Conjugates: Employing Hydrophobic Interactions and DNA Base-Pairing To Create a Diverse Structural Space
A. K. Mishra, H. Weissman, E. Krieg, K. Vatow, M. McCullagh, G. Rybtchinski, F. D. Lewis (2017)
Chemistry: A European Journal, 23, 43, 10328-10337
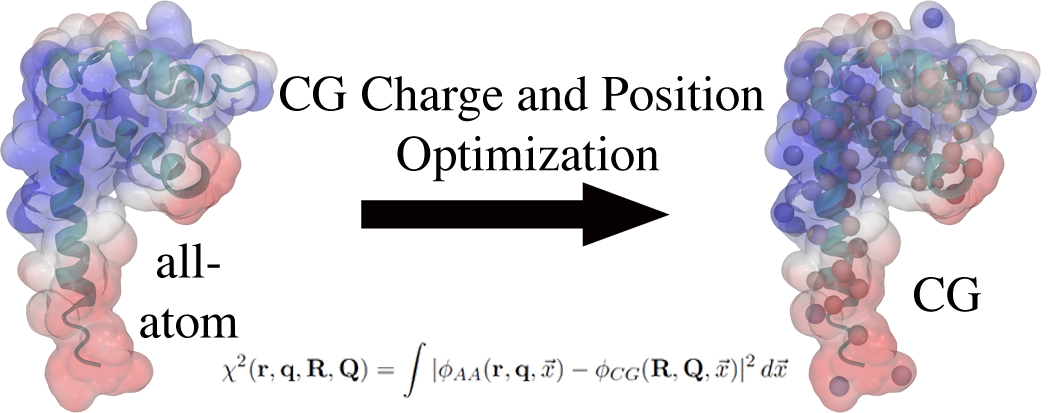 Deriving Coarse-Grained Charges from All-Atom Systems: An Analytic Solution
Deriving Coarse-Grained Charges from All-Atom Systems: An Analytic Solution
P McCullagh, PT Lake, M McCullagh (2016)
J. Chem. Theory Comput., 12, 9, 4390-4399